Difference between revisions of "Glioblastoma"
Jump to navigation
Jump to search
Jensflorian (talk | contribs) (→Microscopic: fix ref) |
Jensflorian (talk | contribs) (→IHC: moved from neuropathology tumours) |
||
| (9 intermediate revisions by the same user not shown) | |||
| Line 34: | Line 34: | ||
*Median survival is measured in months.<ref>{{Cite journal | last1 = Jubelirer | first1 = SJ. | title = A review of the treatment and survival rates of 138 patients with glioblastoma multiforme. | journal = W V Med J | volume = 92 | issue = 4 | pages = 186-90 | month = | year = | doi = | PMID = 8772403 }}</ref> | *Median survival is measured in months.<ref>{{Cite journal | last1 = Jubelirer | first1 = SJ. | title = A review of the treatment and survival rates of 138 patients with glioblastoma multiforme. | journal = W V Med J | volume = 92 | issue = 4 | pages = 186-90 | month = | year = | doi = | PMID = 8772403 }}</ref> | ||
*Only about 5% can expect to survive more than three years.<ref name=pmid17785346>{{Cite journal | last1 = Krex | first1 = D. | last2 = Klink | first2 = B. | last3 = Hartmann | first3 = C. | last4 = von Deimling | first4 = A. | last5 = Pietsch | first5 = T. | last6 = Simon | first6 = M. | last7 = Sabel | first7 = M. | last8 = Steinbach | first8 = JP. | last9 = Heese | first9 = O. | title = Long-term survival with glioblastoma multiforme. | journal = Brain | volume = 130 | issue = Pt 10 | pages = 2596-606 | month = Oct | year = 2007 | doi = 10.1093/brain/awm204 | PMID = 17785346 }}</ref> | *Only about 5% can expect to survive more than three years.<ref name=pmid17785346>{{Cite journal | last1 = Krex | first1 = D. | last2 = Klink | first2 = B. | last3 = Hartmann | first3 = C. | last4 = von Deimling | first4 = A. | last5 = Pietsch | first5 = T. | last6 = Simon | first6 = M. | last7 = Sabel | first7 = M. | last8 = Steinbach | first8 = JP. | last9 = Heese | first9 = O. | title = Long-term survival with glioblastoma multiforme. | journal = Brain | volume = 130 | issue = Pt 10 | pages = 2596-606 | month = Oct | year = 2007 | doi = 10.1093/brain/awm204 | PMID = 17785346 }}</ref> | ||
*Current classification recognizes three types: | |||
** Glioblastoma, IDH-wildtype (aka primary GBM, ICD-O: 9440/3). | |||
** Glioblastoma, IDH-mutant (aka secondary GBM, ICD-O: 9445/3). | |||
** Glioblastoma, NOS (lack of molecular data). | |||
==Macroscopy== | ==Macroscopy== | ||
| Line 116: | Line 120: | ||
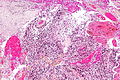
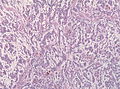
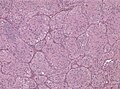
File:GBM layers.jpg | Resection borders in a recurrent GBM (WC/jensflorian) | File:GBM layers.jpg | Resection borders in a recurrent GBM (WC/jensflorian) | ||
File:Glioblastoma-radiation_changes_HE.jpg | Radiation changes in a recurrent GBM (WC/jensflorian) | File:Glioblastoma-radiation_changes_HE.jpg | Radiation changes in a recurrent GBM (WC/jensflorian) | ||
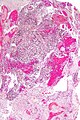
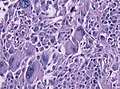
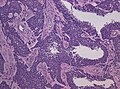
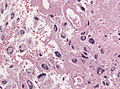
File:GBM_nested_epithelial.jpg | Nested epitheloid appearance in a GBM specimen (WC/jensflorian) | |||
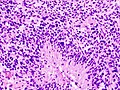
Image:Glioblastoma_%281%29.jpg | Glioblastoma - pseudopalisading of tumour cells (WC) | |||
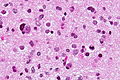
Image:Glioblastoma_-_high_mag.jpg | Glioblastoma with fragment of near-normal white matter - high mag. (WC) | |||
Image:Anaplastic_astrocytoma_-_very_high_mag_-_cropped.jpg | Anaplastic astrocytoma - very high mag. (WC) | |||
</gallery> | </gallery> | ||
www: | www: | ||
| Line 126: | Line 136: | ||
*[[MAP2]] +ve. | *[[MAP2]] +ve. | ||
*[[IDH-1]] -ve (95%). | *[[IDH-1]] -ve (95%). | ||
**+ve if developed from lower grade astrocytoma (secondary GBM). | **+ve if developed from lower grade astrocytoma (secondary GBM) -> classify tumor as Glioblastoma, IDH-mutant. | ||
*[[WT-1]] +ve (cytoplasm). | *[[WT-1]] +ve (cytoplasm). | ||
*p53 +ve (70%). | *p53 +ve (70%). | ||
| Line 133: | Line 143: | ||
*panCK -ve (except for GBM with epithelial component). | *panCK -ve (except for GBM with epithelial component). | ||
*[[ATRX]]: +ve (no loss, nuclei) | *[[ATRX]]: +ve (no loss, nuclei) | ||
**-ve if developed from lower grade astrocytoma (secondary GBM). | **-ve if developed from lower grade astrocytoma (secondary GBM). | ||
*EMA: Dot-like expression less common than in [[ependymoma]]s. | *EMA: Dot-like expression less common than in [[ependymoma]]s. | ||
*MIB-1 usu. 15-30%, but varies greatly. | *MIB-1 usu. 15-30%, but varies greatly. | ||
| Line 141: | Line 151: | ||
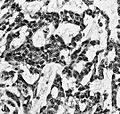
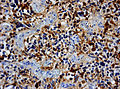
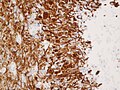
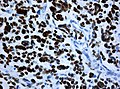
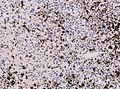
File:Wilms tumor protein wt1 immunohistocehmistry glioblastoma.JPG | WT1 immunostaining in GBM (WC/jensflorian) | File:Wilms tumor protein wt1 immunohistocehmistry glioblastoma.JPG | WT1 immunostaining in GBM (WC/jensflorian) | ||
File:Glioblastoma P53.jpg | GBM with strong p53 immunreactivity (WC/jensflorian) | File:Glioblastoma P53.jpg | GBM with strong p53 immunreactivity (WC/jensflorian) | ||
File:Glioblastoma Ck7.jpg|CK7 staining in glioblastoma with epithelial component (WC/jensflorian) | File:Glioblastoma Ck7.jpg|CK7 staining in glioblastoma with epithelial component (WC/jensflorian) | ||
File:GBM_with_Neuropil_island_MIB1.jpg|Reduced proliferation in neuropil-like islands (MIB1). | File:GBM_with_Neuropil_island_MIB1.jpg|Reduced proliferation in neuropil-like islands (MIB1). | ||
File:GBM_with_Neuropil_island_Syn.jpg |Synaptophysin immunoreactivity in neuropil-like islands (WC/jensflorian). | File:GBM_with_Neuropil_island_Syn.jpg |Synaptophysin immunoreactivity in neuropil-like islands (WC/jensflorian). | ||
Image:Anaplastic_astrocytoma_-_gfap_-_very_high_mag.jpg | Anaplastic astrocytic tumor cells - GFAP - very high mag. (WC) | |||
</gallery> | </gallery> | ||
==Molecular== | ==Molecular== | ||
*IDH1/2 sequencing in cases below 55 years is mandatory to separate between Glioblastoma, IDH-wildtype and Glioblastoma IDH-mutant. | |||
** In cases above 55 years, negative IDH1 R132H immunohistochemistry may be sufficient. | |||
* 70% of IDH-wildtype glioblastoma show chr.7 gain and chr.10 loss.<ref>{{Cite journal | last1 = Ceccarelli | first1 = M. | last2 = Barthel | first2 = FP. | last3 = Malta | first3 = TM. | last4 = Sabedot | first4 = TS. | last5 = Salama | first5 = SR. | last6 = Murray | first6 = BA. | last7 = Morozova | first7 = O. | last8 = Newton | first8 = Y. | last9 = Radenbaugh | first9 = A. | title = Molecular Profiling Reveals Biologically Discrete Subsets and Pathways of Progression in Diffuse Glioma. | journal = Cell | volume = 164 | issue = 3 | pages = 550-63 | month = Jan | year = 2016 | doi = 10.1016/j.cell.2015.12.028 | PMID = 26824661 }}</ref> | |||
*Seen in inherited tumor syndromes: | *Seen in inherited tumor syndromes: | ||
**Lynch | **[[Lynch syndrome]] | ||
**[[Neurofibromatosis 1]] | **[[Neurofibromatosis 1]] | ||
**Li-Fraumeni | **[[Li-Fraumeni syndrome]] | ||
**Turcot-Syndrome | **Turcot-Syndrome | ||
| Line 162: | Line 177: | ||
**PI3K-pathway genes (7-10%). | **PI3K-pathway genes (7-10%). | ||
*Pediatric glioblastoma | |||
**are morphologically indistinct from adult GBM. | |||
**show frequent H3F3A mutations and PDGFRA mutations (Note: H3F3A K27M mutations are classified as [[Astrocytoma#H3.3_K27M_mutated_glioma_of_the_midline|K27 mutated midline glioma]]). | |||
**Consider Epithelioid GBM, when BRAF V600E mutated. | |||
*Diagnostic/therapeutic relevant markers: | *Diagnostic/therapeutic relevant markers: | ||
**MGMT promoter methylation status | **[[MGMT]] promoter methylation status<ref>{{Cite journal | last1 = Quillien | first1 = V. | last2 = Lavenu | first2 = A. | last3 = Karayan-Tapon | first3 = L. | last4 = Carpentier | first4 = C. | last5 = Labussière | first5 = M. | last6 = Lesimple | first6 = T. | last7 = Chinot | first7 = O. | last8 = Wager | first8 = M. | last9 = Honnorat | first9 = J. | title = Comparative assessment of 5 methods (methylation-specific polymerase chain reaction, MethyLight, pyrosequencing, methylation-sensitive high-resolution melting, and immunohistochemistry) to analyze O6-methylguanine-DNA-methyltranferase in a series of 100 glioblastoma patients. | journal = Cancer | volume = 118 | issue = 17 | pages = 4201-11 | month = Sep | year = 2012 | doi = 10.1002/cncr.27392 | PMID = 22294349 }}</ref> | ||
**Absence of LOH 1p/19q (otherwise classify tumor as [[oligodendroglioma]]). | **Absence of LOH 1p/19q (otherwise classify tumor as [[oligodendroglioma]]).<ref>{{Cite journal | last1 = Masui | first1 = K. | last2 = Mischel | first2 = PS. | last3 = Reifenberger | first3 = G. | title = Molecular classification of gliomas. | journal = Handb Clin Neurol | volume = 134 | issue = | pages = 97-120 | month = | year = 2016 | doi = 10.1016/B978-0-12-802997-8.00006-2 | PMID = 26948350 }}</ref> | ||
==See also== | ==See also== | ||
Latest revision as of 09:07, 14 April 2022
| Glioblastoma | |
|---|---|
| Diagnosis in short | |
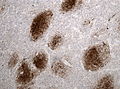
 Glioblastoma. H&E stain. | |
|
| |
| LM | astrocytic differentiation, nuclear atypia, necrosis, microvascular proliferation, +/-pseudopalisading necrosis |
| Subtypes | gliosarcoma |
| LM DDx | anaplastic astrocytoma |
| IHC | GFAP +ve, IDH-1 -ve/+ve |
| Site | brain, spinal cord |
|
| |
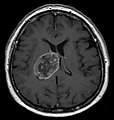
| Radiology | intra-axial |
| Prognosis | very poor |
| Clin. DDx | metastatic brain tumours |
Glioblastoma a very common malignant primary brain tumour in adults. It has a very poor prognosis.
It was previously known as glioblastoma multiforme, abbreviated GBM.
General
- Median survival is measured in months.[1]
- Only about 5% can expect to survive more than three years.[2]
- Current classification recognizes three types:
- Glioblastoma, IDH-wildtype (aka primary GBM, ICD-O: 9440/3).
- Glioblastoma, IDH-mutant (aka secondary GBM, ICD-O: 9445/3).
- Glioblastoma, NOS (lack of molecular data).
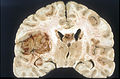
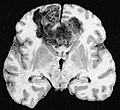
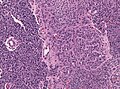
Macroscopy
Features:
- Usu. in white matter.
- central necrotic core.
- ill-defined borders.
- yellowish to dark-brown changes.
- midline shift due to tumor mass.
- In the corpus callosum as bihemispheric "butterfly glioma"
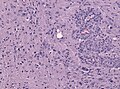
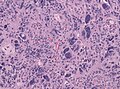
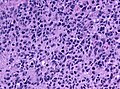
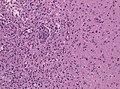
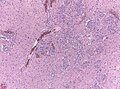
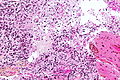
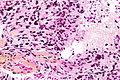
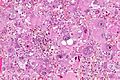
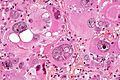
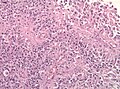
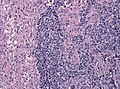
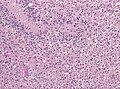
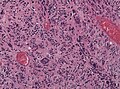
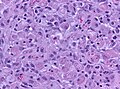
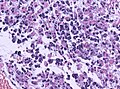
Microscopic
Features:
- Astrocytic tumour with:
- Nuclear atypia.
- Necrosis.
- Endothelial proliferation (AKA microvascular proliferation).
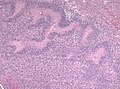
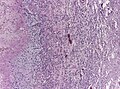
- +/-"Pseudopalisading necrosis" - tumour cells lined-up like a picket fence around necrotic areas.
Glioblastoma variants:
- Giant cell glioblastoma (ICD-O: 9441/3)
- Bizarre multinucleated giant cells.
- Reticulin may be abundant.
- Mean age 44 years, outcome somewhat better than conventional GBM.
- IDH-wildtype, but frequent p53 mutations.
- Epitheloid glioblastoma (ICD-O: 9440/3) [3]
- Closely packed epithelioid to rhabdoid cells, often dicohesive.
- Xanthomaous changes less common than in PXA.
- Children and young adults, outcome particularly poor.
- Up to 50% BRAF V600E mutations.
- Gliosarcoma (ICD-O: 9442/3)
Morphological patterns in Glioblastoma:
- Lipidized (foamy)cells [4]
- Adipocyte-like maturation [5]
- Rhabdoid glioblastoma (focal loss of INI-1) [6]
- Melanotic glioblastoma [7]
- Glioblastoma with oligodendroglial component (no improved survival) [8]
- Granular cell Glioblastoma [9]
- Glioblastoma with primitive neuronal component.[10]
- formerly known as: PNET-like component.
- have a tendency to CSF dissemination.[11]
- Small cell glioblastoma.
- Ependymal-like growth patterns.
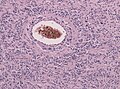
- Glioneuronal tumor with neuropil-like islands.[12]
Images
www:
- Microvascular proliferation in a GBM (ouhsc.edu).
- Pseudopalisading necrosis in GBM (aacrjournals.org).
IHC
- GFAP +ve (cytoplasm).
- MAP2 +ve.
- IDH-1 -ve (95%).
- +ve if developed from lower grade astrocytoma (secondary GBM) -> classify tumor as Glioblastoma, IDH-mutant.
- WT-1 +ve (cytoplasm).
- p53 +ve (70%).
- Neurofilament -ve.
- Synaptophysin -ve (residual Cortex may be +ve).
- panCK -ve (except for GBM with epithelial component).
- ATRX: +ve (no loss, nuclei)
- -ve if developed from lower grade astrocytoma (secondary GBM).
- EMA: Dot-like expression less common than in ependymomas.
- MIB-1 usu. 15-30%, but varies greatly.
Molecular
- IDH1/2 sequencing in cases below 55 years is mandatory to separate between Glioblastoma, IDH-wildtype and Glioblastoma IDH-mutant.
- In cases above 55 years, negative IDH1 R132H immunohistochemistry may be sufficient.
- 70% of IDH-wildtype glioblastoma show chr.7 gain and chr.10 loss.[13]
- Seen in inherited tumor syndromes:
- Lynch syndrome
- Neurofibromatosis 1
- Li-Fraumeni syndrome
- Turcot-Syndrome
- Most common alterations (TCGA[14])
- Tp53 (42% of the tumors mutated)
- PTEN (33%).
- NF1 (21%).
- EGFR (18%).
- RB1 (11%).
- PI3K-pathway genes (7-10%).
- Pediatric glioblastoma
- are morphologically indistinct from adult GBM.
- show frequent H3F3A mutations and PDGFRA mutations (Note: H3F3A K27M mutations are classified as K27 mutated midline glioma).
- Consider Epithelioid GBM, when BRAF V600E mutated.
- Diagnostic/therapeutic relevant markers:
- MGMT promoter methylation status[15]
- Absence of LOH 1p/19q (otherwise classify tumor as oligodendroglioma).[16]
See also
References
- ↑ Jubelirer, SJ.. "A review of the treatment and survival rates of 138 patients with glioblastoma multiforme.". W V Med J 92 (4): 186-90. PMID 8772403.
- ↑ Krex, D.; Klink, B.; Hartmann, C.; von Deimling, A.; Pietsch, T.; Simon, M.; Sabel, M.; Steinbach, JP. et al. (Oct 2007). "Long-term survival with glioblastoma multiforme.". Brain 130 (Pt 10): 2596-606. doi:10.1093/brain/awm204. PMID 17785346.
- ↑ te journal | last1 = Kleinschmidt-DeMasters | first1 = BK. | last2 = Aisner | first2 = DL. | last3 = Birks | first3 = DK. | last4 = Foreman | first4 = NK. | title = Epithelioid GBMs show a high percentage of BRAF V600E mutation. | journal = Am J Surg Pathol | volume = 37 | issue = 5 | pages = 685-98 | month = May | year = 2013 | doi = 10.1097/PAS.0b013e31827f9c5e | PMID = 23552385 }}
- ↑ Kepes, JJ.; Rubinstein, LJ. (May 1981). "Malignant gliomas with heavily lipidized (foamy) tumor cells: a report of three cases with immunoperoxidase study.". Cancer 47 (10): 2451-9. PMID 7023643.
- ↑ Rickert, CH.; Riemenschneider, MJ.; Schachenmayr, W.; Richter, HP.; Bockhorn, J.; Reifenberger, G.; Paulus, W. (Jul 2009). "Glioblastoma with adipocyte-like tumor cell differentiation--histological and molecular features of a rare differentiation pattern.". Brain Pathol 19 (3): 431-8. doi:10.1111/j.1750-3639.2008.00199.x. PMID 18691268.
- ↑ Hiroyuki, M.; Ogino, J.; Takahashi, A.; Hasegawa, T.; Wakabayashi, T. (Feb 2015). "Rhabdoid glioblastoma: an aggressive variaty of astrocytic tumor.". Nagoya J Med Sci 77 (1-2): 321-8. PMID 25797998.
- ↑ Jaiswal, S.; Agrawal, V.; Vij, M.; Sahu, RN.; Jaiswal, AK.; Behari, S.. "Glioblastoma with melanotic differentiation.". Clin Neuropathol 29 (5): 330-3. PMID 20860897.
- ↑ Hegi, ME.; Janzer, RC.; Lambiv, WL.; Gorlia, T.; Kouwenhoven, MC.; Hartmann, C.; von Deimling, A.; Martinet, D. et al. (Jun 2012). "Presence of an oligodendroglioma-like component in newly diagnosed glioblastoma identifies a pathogenetically heterogeneous subgroup and lacks prognostic value: central pathology review of the EORTC_26981/NCIC_CE.3 trial.". Acta Neuropathol 123 (6): 841-52. doi:10.1007/s00401-011-0938-4. PMID 22249618.
- ↑ Schittenhelm, J.; Psaras, T.. "Glioblastoma with granular cell astrocytoma features: a case report and literature review.". Clin Neuropathol 29 (5): 323-9. PMID 20860896.
- ↑ Louis, DN.; Perry, A.; Reifenberger, G.; von Deimling, A.; Figarella-Branger, D.; Cavenee, WK.; Ohgaki, H.; Wiestler, OD. et al. (Jun 2016). "The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary.". Acta Neuropathol 131 (6): 803-20. doi:10.1007/s00401-016-1545-1. PMID 27157931.
- ↑ Perry, A.; Miller, CR.; Gujrati, M.; Scheithauer, BW.; Zambrano, SC.; Jost, SC.; Raghavan, R.; Qian, J. et al. (Jan 2009). "Malignant gliomas with primitive neuroectodermal tumor-like components: a clinicopathologic and genetic study of 53 cases.". Brain Pathol 19 (1): 81-90. doi:10.1111/j.1750-3639.2008.00167.x. PMID 18452568.
- ↑ Ishizawa, K.; Hirose, T.; Sugiyama, K.; Kageji, T.; Nobusawa, S.; Homma, T.; Komori, T.; Sasaki, A.. "Pathologic diversity of glioneuronal tumor with neuropil-like islands: a histological and immunohistochemical study with a special reference to isocitrate dehydrogenase 1 (IDH1) in 5 cases.". Clin Neuropathol 31 (2): 67-76. PMID 22385787.
- ↑ Ceccarelli, M.; Barthel, FP.; Malta, TM.; Sabedot, TS.; Salama, SR.; Murray, BA.; Morozova, O.; Newton, Y. et al. (Jan 2016). "Molecular Profiling Reveals Biologically Discrete Subsets and Pathways of Progression in Diffuse Glioma.". Cell 164 (3): 550-63. doi:10.1016/j.cell.2015.12.028. PMID 26824661.
- ↑ Verhaak, RG.; Hoadley, KA.; Purdom, E.; Wang, V.; Qi, Y.; Wilkerson, MD.; Miller, CR.; Ding, L. et al. (Jan 2010). "Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1.". Cancer Cell 17 (1): 98-110. doi:10.1016/j.ccr.2009.12.020. PMID 20129251.
- ↑ Quillien, V.; Lavenu, A.; Karayan-Tapon, L.; Carpentier, C.; Labussière, M.; Lesimple, T.; Chinot, O.; Wager, M. et al. (Sep 2012). "Comparative assessment of 5 methods (methylation-specific polymerase chain reaction, MethyLight, pyrosequencing, methylation-sensitive high-resolution melting, and immunohistochemistry) to analyze O6-methylguanine-DNA-methyltranferase in a series of 100 glioblastoma patients.". Cancer 118 (17): 4201-11. doi:10.1002/cncr.27392. PMID 22294349.
- ↑ Masui, K.; Mischel, PS.; Reifenberger, G. (2016). "Molecular classification of gliomas.". Handb Clin Neurol 134: 97-120. doi:10.1016/B978-0-12-802997-8.00006-2. PMID 26948350.