Difference between revisions of "Medulloblastoma"
Jump to navigation
Jump to search
Jensflorian (talk | contribs) |
Jensflorian (talk | contribs) |
||
| (One intermediate revision by the same user not shown) | |||
| Line 1: | Line 1: | ||
{{ Infobox diagnosis | |||
| Name = {{PAGENAME}} | |||
| Image = Medulloblastoma with rosettes.jpg | |||
| Width = | |||
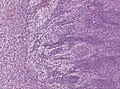
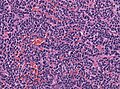
| Caption = Classic medulloblastoma [[H&E stain]]. | |||
| Synonyms = | |||
| Micro = | |||
| Subtypes = | |||
| LMDDx = small cell round blue tumous. | |||
| Stains = Reticulin +ve (in desmoplastic MB) | |||
| IHC = MAP2+ve | |||
| EM = | |||
| Molecular = | |||
| IF = | |||
| Gross = cerebellar. | |||
| Grossing = | |||
| Site = brain - usu. [[cerebellum]] | |||
| Assdx = | |||
| Syndromes = | |||
| Clinicalhx = | |||
| Signs = | |||
| Symptoms = | |||
| Prevalence = common - esp. in children | |||
| Bloodwork = | |||
| Rads = | |||
| Endoscopy = | |||
| Prognosis = subtype-dependent (WHO Grade IV) | |||
| Other = | |||
| ClinDDx = | |||
| Tx = | |||
}} | |||
'''Medulloblastoma''' is a [[malignant]] [[small round cell tumour]] that is found in the [[cerebellum]] or dorsal brainstem. It is the most common malignant CNS tumour in children. | '''Medulloblastoma''' is a [[malignant]] [[small round cell tumour]] that is found in the [[cerebellum]] or dorsal brainstem. It is the most common malignant CNS tumour in children. | ||
| Line 118: | Line 150: | ||
**Infantile, p53 wildtype: Usu. desmoplastic/nodular, 10q loss. | **Infantile, p53 wildtype: Usu. desmoplastic/nodular, 10q loss. | ||
**SHH-p53 mutant: Usu. large cell/anaplastic, 17q loss. | **SHH-p53 mutant: Usu. large cell/anaplastic, 17q loss. | ||
*Group 3 (approx. 20%). | *Non WNT/Non SHH: | ||
**Classic and large cell/anaplastic, MYC amplification, isochromosome 17q. | **Group 3 (approx. 20%). | ||
*Group 4 (approx. 40%). | ***Classic and large cell/anaplastic, MYC amplification, isochromosome 17q. | ||
**Classic phenotype, MYCN amplification, isodicentric 17q. | **Group 4 (approx. 40%). | ||
***Classic phenotype, MYCN amplification, isodicentric 17q. | |||
Note: Within Group 3+4 two or more of chromosome 7 gain, chromosome 8 loss, and chromosome 11 loss separates standard risk medulloblastoma samples into favorable and classifies the remaining i17q diploid cases as high-risk. | |||
==Prognosis== | ==Prognosis== | ||
Latest revision as of 15:20, 20 November 2019
| Medulloblastoma | |
|---|---|
| Diagnosis in short | |
 Classic medulloblastoma H&E stain. | |
| LM DDx | small cell round blue tumous. |
| Stains | Reticulin +ve (in desmoplastic MB) |
| IHC | MAP2+ve |
| Gross | cerebellar. |
| Site | brain - usu. cerebellum |
|
| |
| Prevalence | common - esp. in children |
| Prognosis | subtype-dependent (WHO Grade IV) |
Medulloblastoma is a malignant small round cell tumour that is found in the cerebellum or dorsal brainstem. It is the most common malignant CNS tumour in children.
General
- Mostly paediatric population (mean age: 9 years).
- Tumors consists of histologically and molecular distinct subgroups.
- All subgroups correspond to WHO grade IV.
- May be seen as a component of nevoid basal cell carcinoma syndrome (NBCCS) AKA Gorlin syndrome, Li-Fraumeni syndrome, Turcot syndrome, Rubinstain-Taybi syndrome and Nijmegen breakage syndrome.
- Show molecularly spatially homogeneous transcriptomes, allowing for accurate subgrouping of tumors from a single biopsy.[1]
- Commonly spread via cerebrospinal fluid (CSF).[2]
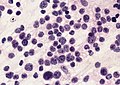
- May be detected in CSF cytopathology specimens.
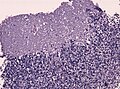
Gross
- Location: cerebellum - key feature or dorsal brainstem (lower rhombic lip).
- For morphologically identical supratentorial tumours see possible DDx.
- Supratentorial and spinal metastases from initial medulloblastoma possible.
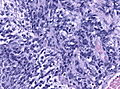
Microscopic
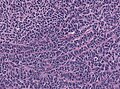
Features:[3]
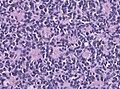
- Small round cell tumour.
- Mild to moderate nuclear pleomorphism.
- High mitotic count.
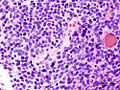
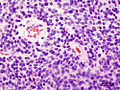
- Homer-Wright rosettes:
- Rosette with a meshwork of fibers (neuropil) at the centre.[4]
IHC
- MAP2 usu. +ve
- Synaptophysin +ve (weak to strong)
- NSE +ve/-ve
- NF +ve/-ve
- Chromogranin +ve/-ve
- GFAP +ve/-ve (mostly along blood vessels)
- Vimentin +ve
- Nestin +ve
- INI1 retained (no loss)
Genetic grouping with IHC:[5]
- WNT: beta-catein: nuclear +ve, YAP1+ve, OTX2+ve.
- SHH: YAP1+ve, OTX2-ve, p75+ve.
- Non-WNT/Non-SHH: * SHH: YAP1-ve, OTX2+ve, p75+ve.
DDx:
Images
Case:
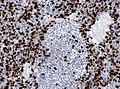
Partial MAP2 immunoreactivity. (WC/jensflorian)
www:
- Medulloblastoma (ouhsc.edu).
- Medulloblastoma - several images (upmc.edu).
- Medulloblastoma with rhabdomyoblastic differentiation - several images (upmc.edu).
Subtypes
Histologically defined
- Classic medulloblastoma (~85% of all medulloblastomas).
- Variants of medulloblastoma (~15% of all medulloblastomas together):
- Anaplastic / Large cell variant.
- Desmoplastic/nodular medulloblastoma (DNMB).
- Medulloblastoma with extensive nodularity (MBEN).
Classic variant
- Densely packed embryonal cells.
- Mitosis / apoptosis present.
- Nodules of neurocytic differentiation without internodular reticulin.
- Rarely spongioblastic pattern (ribboning).
Anaplastic variant
Features:
- Larger cells.
- Severe anaplasia.
- Polygonal cells.
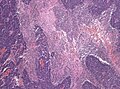
Desmoplastic/nodular variant
Features:
- Nodular, reticulin-free zones (pale islands).
- Mitotic activity is reduced.
- Densely packed internodal, reticulin-positive tumor areas.
- Absence of neuroblastic rosettes.
- Neuronal markers (NeuN, Synaptophysin) can be +ve.
Genetically defined
- WNT (brainstem-related, usu. CTNNB1 mutations, less common: CSNK2B, AXIN2, APC, approx. 10%).[7]
- Usu. classic MB in histology.
- Caution: Some WNT-activated MB may show additional subclonal SHH-mutations.[8]
- Peak incidence: 7-14 years (15% adults).
- Beta-catenin nuclear +ve.
- Excellent prognosis.
- SHH (PTCH1 & SUFU in infant, PTCH1, SMO, Gli2, MYCN in non-infant, approx 30%).[9]
- Origin in the cerebellar granule neuron cell precursors.
- Infantile and adult cases are biologically different, esp p53 mutant tumors are associated with poor outcome.[10]
- Infantile, p53 wildtype: Usu. desmoplastic/nodular, 10q loss.
- SHH-p53 mutant: Usu. large cell/anaplastic, 17q loss.
- Non WNT/Non SHH:
- Group 3 (approx. 20%).
- Classic and large cell/anaplastic, MYC amplification, isochromosome 17q.
- Group 4 (approx. 40%).
- Classic phenotype, MYCN amplification, isodicentric 17q.
- Group 3 (approx. 20%).
Note: Within Group 3+4 two or more of chromosome 7 gain, chromosome 8 loss, and chromosome 11 loss separates standard risk medulloblastoma samples into favorable and classifies the remaining i17q diploid cases as high-risk.
Prognosis
- Prognosis based on histology:[11][12][13] DNMB & MBEN > classic > anaplastic & large cell variant.
- Prognosis based on genetics:[14] WNT > SHH (without Tp53 mut) > Group 4 > Group 3 > SHH (with Tp53 mut).
See also
References
- ↑ Morrissy, AS.; Cavalli, FMG.; Remke, M.; Ramaswamy, V.; Shih, DJH.; Holgado, BL.; Farooq, H.; Donovan, LK. et al. (May 2017). "Spatial heterogeneity in medulloblastoma.". Nat Genet 49 (5): 780-788. doi:10.1038/ng.3838. PMID 28394352.
- ↑ Lefkowitch, Jay H. (2006). Anatomic Pathology Board Review (1st ed.). Saunders. pp. 424 Q34. ISBN 978-1416025887.
- ↑ URL: http://moon.ouhsc.edu/kfung/jty1/neurotest/Q93-Ans.htm. Accessed on: 26 October 2010.
- ↑ Wippold FJ, Perry A (March 2006). "Neuropathology for the neuroradiologist: rosettes and pseudorosettes". AJNR Am J Neuroradiol 27 (3): 488–92. PMID 16551982.
- ↑ Pietsch, T.; Haberler, C.. "Update on the integrated histopathological and genetic classification of medulloblastoma - a practical diagnostic guideline.". Clin Neuropathol 35 (6): 344-352. doi:10.5414/NP300999. PMID 27781424.
- ↑ Phi, JH.; Park, AK.; Lee, S.; Choi, SA.; Baek, IP.; Kim, P.; Kim, EH.; Park, HC. et al. (Apr 2018). "Genomic analysis reveals secondary glioblastoma after radiotherapy in a subset of recurrent medulloblastomas.". Acta Neuropathol. doi:10.1007/s00401-018-1845-8. PMID 29644394.
- ↑ Wefers, AK.; Warmuth-Metz, M.; Pöschl, J.; von Bueren, AO.; Monoranu, CM.; Seelos, K.; Peraud, A.; Tonn, JC. et al. (2014). "Subgroup-specific localization of human medulloblastoma based on pre-operative MRI.". Acta Neuropathol 127 (6): 931-3. doi:10.1007/s00401-014-1271-5. PMID 24699697.
- ↑ Iorgulescu, JB.; Van Ziffle, J.; Stevers, M.; Grenert, JP.; Bastian, BC.; Chavez, L.; Stichel, D.; Buchhalter, I. et al. (Apr 2018). "Deep sequencing of WNT-activated medulloblastomas reveals secondary SHH pathway activation.". Acta Neuropathol 135 (4): 635-638. doi:10.1007/s00401-018-1819-x. PMID 29435664.
- ↑ Neumann, JE.; Swartling, FJ.; Schüller, U. (Jul 2017). "Medulloblastoma: experimental models and reality.". Acta Neuropathol. doi:10.1007/s00401-017-1753-3. PMID 28725965.
- ↑ Kool, M.; Jones, DT.; Jäger, N.; Northcott, PA.; Pugh, TJ.; Hovestadt, V.; Piro, RM.; Esparza, LA. et al. (Mar 2014). "Genome sequencing of SHH medulloblastoma predicts genotype-related response to smoothened inhibition.". Cancer Cell 25 (3): 393-405. doi:10.1016/j.ccr.2014.02.004. PMID 24651015.
- ↑ Gulino A, Arcella A, Giangaspero F (November 2008). "Pathological and molecular heterogeneity of medulloblastoma". Curr Opin Oncol 20 (6): 668–75. doi:10.1097/CCO.0b013e32831369f4. PMID 18841049.
- ↑ Rutkowski S, von Hoff K, Emser A, et al. (November 2010). "Survival and Prognostic Factors of Early Childhood Medulloblastoma: An International Meta-Analysis". J Clin Oncol 28 (33): 4961–4968. doi:10.1200/JCO.2010.30.2299. PMID 20940197.
- ↑ Rutkowski, S.; Bode, U.; Deinlein, F.; Ottensmeier, H.; Warmuth-Metz, M.; Soerensen, N.; Graf, N.; Emser, A. et al. (Mar 2005). "Treatment of early childhood medulloblastoma by postoperative chemotherapy alone.". N Engl J Med 352 (10): 978-86. doi:10.1056/NEJMoa042176. PMID 15758008.
- ↑ Ramaswamy, V.; Remke, M.; Bouffet, E.; Bailey, S.; Clifford, SC.; Doz, F.; Kool, M.; Dufour, C. et al. (Jun 2016). "Risk stratification of childhood medulloblastoma in the molecular era: the current consensus.". Acta Neuropathol 131 (6): 821-31. doi:10.1007/s00401-016-1569-6. PMID 27040285.