Medulloblastoma
Jump to navigation
Jump to search
The printable version is no longer supported and may have rendering errors. Please update your browser bookmarks and please use the default browser print function instead.
| Medulloblastoma | |
|---|---|
| Diagnosis in short | |
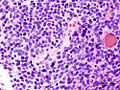
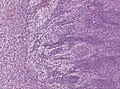
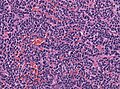
 Classic medulloblastoma H&E stain. | |
| LM DDx | small cell round blue tumous. |
| Stains | Reticulin +ve (in desmoplastic MB) |
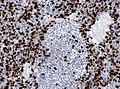
| IHC | MAP2+ve |
| Gross | cerebellar. |
| Site | brain - usu. cerebellum |
|
| |
| Prevalence | common - esp. in children |
| Prognosis | subtype-dependent (WHO Grade IV) |
Medulloblastoma is a malignant small round cell tumour that is found in the cerebellum or dorsal brainstem. It is the most common malignant CNS tumour in children.
General
- Mostly paediatric population (mean age: 9 years).
- Tumors consists of histologically and molecular distinct subgroups.
- All subgroups correspond to WHO grade IV.
- May be seen as a component of nevoid basal cell carcinoma syndrome (NBCCS) AKA Gorlin syndrome, Li-Fraumeni syndrome, Turcot syndrome, Rubinstain-Taybi syndrome and Nijmegen breakage syndrome.
- Show molecularly spatially homogeneous transcriptomes, allowing for accurate subgrouping of tumors from a single biopsy.[1]
- Commonly spread via cerebrospinal fluid (CSF).[2]
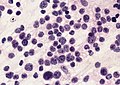
- May be detected in CSF cytopathology specimens.
Gross
- Location: cerebellum - key feature or dorsal brainstem (lower rhombic lip).
- For morphologically identical supratentorial tumours see possible DDx.
- Supratentorial and spinal metastases from initial medulloblastoma possible.
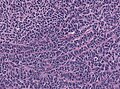
Microscopic
Features:[3]
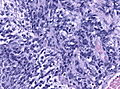
- Small round cell tumour.
- Mild to moderate nuclear pleomorphism.
- High mitotic count.
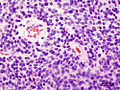
- Homer-Wright rosettes:
- Rosette with a meshwork of fibers (neuropil) at the centre.[4]
IHC
- MAP2 usu. +ve
- Synaptophysin +ve (weak to strong)
- NSE +ve/-ve
- NF +ve/-ve
- Chromogranin +ve/-ve
- GFAP +ve/-ve (mostly along blood vessels)
- Vimentin +ve
- Nestin +ve
- INI1 retained (no loss)
Genetic grouping with IHC:[5]
- WNT: beta-catein: nuclear +ve, YAP1+ve, OTX2+ve.
- SHH: YAP1+ve, OTX2-ve, p75+ve.
- Non-WNT/Non-SHH: * SHH: YAP1-ve, OTX2+ve, p75+ve.
DDx:
Images
Case:
Partial MAP2 immunoreactivity. (WC/jensflorian)
www:
- Medulloblastoma (ouhsc.edu).
- Medulloblastoma - several images (upmc.edu).
- Medulloblastoma with rhabdomyoblastic differentiation - several images (upmc.edu).
Subtypes
Histologically defined
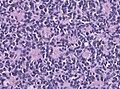
- Classic medulloblastoma (~85% of all medulloblastomas).
- Variants of medulloblastoma (~15% of all medulloblastomas together):
- Anaplastic / Large cell variant.
- Desmoplastic/nodular medulloblastoma (DNMB).
- Medulloblastoma with extensive nodularity (MBEN).
Classic variant
- Densely packed embryonal cells.
- Mitosis / apoptosis present.
- Nodules of neurocytic differentiation without internodular reticulin.
- Rarely spongioblastic pattern (ribboning).
Anaplastic variant
Features:
- Larger cells.
- Severe anaplasia.
- Polygonal cells.
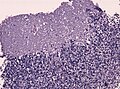
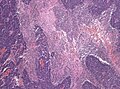
Desmoplastic/nodular variant
Features:
- Nodular, reticulin-free zones (pale islands).
- Mitotic activity is reduced.
- Densely packed internodal, reticulin-positive tumor areas.
- Absence of neuroblastic rosettes.
- Neuronal markers (NeuN, Synaptophysin) can be +ve.
Genetically defined
- WNT (brainstem-related, usu. CTNNB1 mutations, less common: CSNK2B, AXIN2, APC, approx. 10%).[7]
- Usu. classic MB in histology.
- Caution: Some WNT-activated MB may show additional subclonal SHH-mutations.[8]
- Peak incidence: 7-14 years (15% adults).
- Beta-catenin nuclear +ve.
- Excellent prognosis.
- SHH (PTCH1 & SUFU in infant, PTCH1, SMO, Gli2, MYCN in non-infant, approx 30%).[9]
- Origin in the cerebellar granule neuron cell precursors.
- Infantile and adult cases are biologically different, esp p53 mutant tumors are associated with poor outcome.[10]
- Infantile, p53 wildtype: Usu. desmoplastic/nodular, 10q loss.
- SHH-p53 mutant: Usu. large cell/anaplastic, 17q loss.
- Non WNT/Non SHH:
- Group 3 (approx. 20%).
- Classic and large cell/anaplastic, MYC amplification, isochromosome 17q.
- Group 4 (approx. 40%).
- Classic phenotype, MYCN amplification, isodicentric 17q.
- Group 3 (approx. 20%).
Note: Within Group 3+4 two or more of chromosome 7 gain, chromosome 8 loss, and chromosome 11 loss separates standard risk medulloblastoma samples into favorable and classifies the remaining i17q diploid cases as high-risk.
Prognosis
- Prognosis based on histology:[11][12][13] DNMB & MBEN > classic > anaplastic & large cell variant.
- Prognosis based on genetics:[14] WNT > SHH (without Tp53 mut) > Group 4 > Group 3 > SHH (with Tp53 mut).
See also
References
- ↑ Morrissy, AS.; Cavalli, FMG.; Remke, M.; Ramaswamy, V.; Shih, DJH.; Holgado, BL.; Farooq, H.; Donovan, LK. et al. (May 2017). "Spatial heterogeneity in medulloblastoma.". Nat Genet 49 (5): 780-788. doi:10.1038/ng.3838. PMID 28394352.
- ↑ Lefkowitch, Jay H. (2006). Anatomic Pathology Board Review (1st ed.). Saunders. pp. 424 Q34. ISBN 978-1416025887.
- ↑ URL: http://moon.ouhsc.edu/kfung/jty1/neurotest/Q93-Ans.htm. Accessed on: 26 October 2010.
- ↑ Wippold FJ, Perry A (March 2006). "Neuropathology for the neuroradiologist: rosettes and pseudorosettes". AJNR Am J Neuroradiol 27 (3): 488–92. PMID 16551982.
- ↑ Pietsch, T.; Haberler, C.. "Update on the integrated histopathological and genetic classification of medulloblastoma - a practical diagnostic guideline.". Clin Neuropathol 35 (6): 344-352. doi:10.5414/NP300999. PMID 27781424.
- ↑ Phi, JH.; Park, AK.; Lee, S.; Choi, SA.; Baek, IP.; Kim, P.; Kim, EH.; Park, HC. et al. (Apr 2018). "Genomic analysis reveals secondary glioblastoma after radiotherapy in a subset of recurrent medulloblastomas.". Acta Neuropathol. doi:10.1007/s00401-018-1845-8. PMID 29644394.
- ↑ Wefers, AK.; Warmuth-Metz, M.; Pöschl, J.; von Bueren, AO.; Monoranu, CM.; Seelos, K.; Peraud, A.; Tonn, JC. et al. (2014). "Subgroup-specific localization of human medulloblastoma based on pre-operative MRI.". Acta Neuropathol 127 (6): 931-3. doi:10.1007/s00401-014-1271-5. PMID 24699697.
- ↑ Iorgulescu, JB.; Van Ziffle, J.; Stevers, M.; Grenert, JP.; Bastian, BC.; Chavez, L.; Stichel, D.; Buchhalter, I. et al. (Apr 2018). "Deep sequencing of WNT-activated medulloblastomas reveals secondary SHH pathway activation.". Acta Neuropathol 135 (4): 635-638. doi:10.1007/s00401-018-1819-x. PMID 29435664.
- ↑ Neumann, JE.; Swartling, FJ.; Schüller, U. (Jul 2017). "Medulloblastoma: experimental models and reality.". Acta Neuropathol. doi:10.1007/s00401-017-1753-3. PMID 28725965.
- ↑ Kool, M.; Jones, DT.; Jäger, N.; Northcott, PA.; Pugh, TJ.; Hovestadt, V.; Piro, RM.; Esparza, LA. et al. (Mar 2014). "Genome sequencing of SHH medulloblastoma predicts genotype-related response to smoothened inhibition.". Cancer Cell 25 (3): 393-405. doi:10.1016/j.ccr.2014.02.004. PMID 24651015.
- ↑ Gulino A, Arcella A, Giangaspero F (November 2008). "Pathological and molecular heterogeneity of medulloblastoma". Curr Opin Oncol 20 (6): 668–75. doi:10.1097/CCO.0b013e32831369f4. PMID 18841049.
- ↑ Rutkowski S, von Hoff K, Emser A, et al. (November 2010). "Survival and Prognostic Factors of Early Childhood Medulloblastoma: An International Meta-Analysis". J Clin Oncol 28 (33): 4961–4968. doi:10.1200/JCO.2010.30.2299. PMID 20940197.
- ↑ Rutkowski, S.; Bode, U.; Deinlein, F.; Ottensmeier, H.; Warmuth-Metz, M.; Soerensen, N.; Graf, N.; Emser, A. et al. (Mar 2005). "Treatment of early childhood medulloblastoma by postoperative chemotherapy alone.". N Engl J Med 352 (10): 978-86. doi:10.1056/NEJMoa042176. PMID 15758008.
- ↑ Ramaswamy, V.; Remke, M.; Bouffet, E.; Bailey, S.; Clifford, SC.; Doz, F.; Kool, M.; Dufour, C. et al. (Jun 2016). "Risk stratification of childhood medulloblastoma in the molecular era: the current consensus.". Acta Neuropathol 131 (6): 821-31. doi:10.1007/s00401-016-1569-6. PMID 27040285.